Brain Region Assignment
The original 16,050 reconstructions in the Fly Circuit database (flycircuit.tw) were associated with the somatic coordinates of each neuron within the three-dimensional framework of male and female brain templates. Both templates parcel the fly brain in 58 regions (29 in each hemisphere). However, the regional assignment for the somata of the reconstructed neurons was not provided. To mirror this dataset in NeuroMorpho.Org (v6.0) we computed the somatic region from the available metadata, brain image stacks, and query tools present in the flycircuit.tw website. This document explains our method of mapping each neuron to NeuroMorpho.Org brain region metadata.
We first used the text-based query tool in flycircuit.tw to search for neurons present within 0 micrometers of each region. This operation returned 5533 neurons mapped to single regions and no neuron mapped to more than one region. Then we searched neurons within 10 microns of each region. This step returned 8171 neurons mapped to single regions, including 3958 of the 5533 found at 0 microns. The brain region matched between 0 and 10 microns for 3894 of those 3958 neurons, and mismatched for only 64. Assuming the assignment at 0 microns as the gold standard, these values correspond to 98.4% reliability for the assignments at 10 microns. We therefore accepted the further assignment of the additional (8171-3958=) 4213 neurons. With the same approach, we further assigned the somatic brain region of 2475 more neurons (with estimated 84.2% reliability) by searching within 20 microns. When we attempted to search within 30 microns, however, the estimated reliability dropped to 13.6%. Thus, we rejected those assignments. Overall, the above described process assigned the somatic brain region of (5533+4213+2475=) 12,219 neurons.
The search within 10 microns also returned 1984 neurons matching two or more brain regions. Similarly, the search within 20 microns also returned 535 additional neurons matching two or more brain regions. Lastly, 1489 neurons could not be mapped to any region within 20 microns and were considered as potentially residing in any of the 58 regions. Note that the total sums up to 16,227, but only 16,050 of these neurons had an associated morphological reconstruction file.
The somatic assignment of every neuron matching two or more brain regions proceeded as the following. We first calculated the Euclidean distance between that neuron and each of the neurons uniquely mapped to one of those regions (a subset of the 12,219 uniquely mapped neurons). We then defined the proximity of the neuron to each of those regions as the sum of the inverse of the distance to every neuron in that region. For example, if a neuron matches both Mushroom Body (MB) and Medulla (Med) in one hemisphere, given the distances of that neuron from the n Mushroom Body neurons (dMB1, dMB2,dMB3,.....dMBn) and from the m Medulla neurons (dMed1, dMed2, dMed3, dMed3,.....dMedm), the respective proximities are:
ProxMB = Σi(1/dMBi), where i=1...n, and ProxMed = Σj(1/dMedj), where j=1...m.
These proximity values are then used to determine the relative assignment probabilities for MB and MED:
ProbMB = ProxMB / (ProxMB + ProxMed) and ProbMed = ProxMed / (ProxMB + ProxMed).
The somata of fly neurons can belong to one region or lie on the border between two, or occasionally three, adjacent regions. If a neuron only matched two regions, we assigned the somatic location exclusively to one region if its probability was at least twice as high as that of the other region (that is, if one of the two regions had a probability of 66.67% or higher). Otherwise, we assigned the neuron to both regions (representing a border location between the two). For neurons matching three or more region, we devised a decision tree to associate one, two or three regions (representing their location in one region, in the border of two regions, and in the intersection of three regions respectively) based on the relative assignment probabilities P1, P2, and P3 of the regions with top proximities. The decision used a probability threshold of 2/(k+1), where k is the number of matching regions (k = 3...8 or 58) . For example, the threshold is 50% for neurons matching three regions, 22.2% for neurons matching 8 regions, and 3.39% for neurons matching all 58 regions (that is, matching none of them).
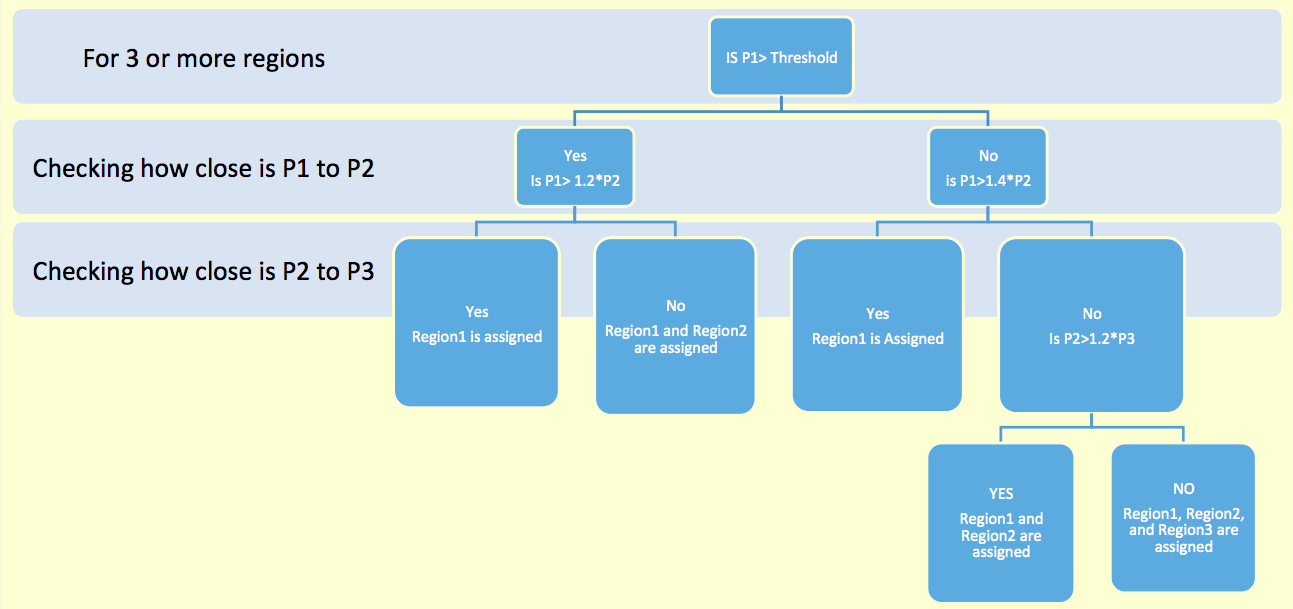
For example, for a neuron matching three regions, if the probability of the most likely region is above threshold (50%), but within 20% of the probability of the second most likely region, then the soma is assigned to both of the top two regions. Alternatively, if the probability of the most likely region is below the 50% threshold, but still above 40% of the probability of the second most likely region, then the soma is only assigned to the top region.
Next we built an adjacency matrix of male and female brain regions based on the corresponding template image stacks from the Fly Circuit. We considered two regions adjacent if they share a border or if they are in close proximity, without a third region in the middle. We employed this matrix to verify that the double or triple somatic region assignments by the decision tree corresponded to adjacent cases. All doubtful cases were manually checked by visualizing the corresponding brain-embedded neuronal reconstruction on the Fly Circuit website.
Lastly, we mapped the assigned regions to appropriate metadata entries in NeuroMorpho.Org based on the Virtual Fly Brain hierarchy (VirtualFlyBrain.org). For neurons assigned to more than one region, we mapped only the most likely region to the Virtual Fly Brain hierarchy, adding the other assigned region(s) at the lowest regional level of NeuroMorpho.Org metadata.
Cell Type Assignment
We use the Fly Circuit metadata and brain region adjacency matrix (described in the Brain Region Assignment section) to divide all reconstructions into principal (projection) cells and interneurons. Specifically, the available metadata included the list of regions that contained neurite terminals for each neuron. A neuron was classified as interneuron if 95% or more of its neurite terminals were contained within the somatic region and its adjacent brain regions. Conversely, a neuron was classified as a principal cell if more than 5% of its neurite terminals were found in non-adjacent regions. This definition yielded 10,079 principal cells and 5,971 interneurons. We further sub-divided all neurons on the basis of their putative neurotransmitter and, as a tertiary hierarchical level, by their birth date.
Citation
Nanda S, Allaham M, Bergamino M, Polavaram S, Armañanzas R, Ascoli GA & Parekh R (2015) Doubling up on the fly: NeuroMorpho.Org meets Big Data. Neuroinformatics, DOI 10.1007/s12021-014-9257-y
|