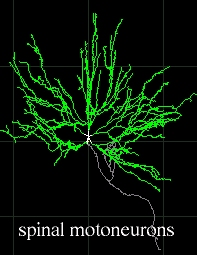
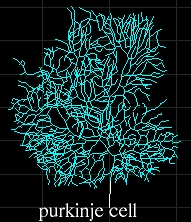
 Examples
of
Neuronal Reconstructions
Examples
of
Neuronal Reconstructions
This tool allows researchers to extract quantitative morphological
measurements from neuronal reconstructions. Neuronal reconstructions
are typically obtained from brightfield or fluorescence microscopy
preparations using applications such as Neurolucida, Eutectic, or
Neuron_Morpho, or can be synthesized via computational simulations.
Several hundreds neuronal reconstructions are freely available to the
neuroscience community (via web archives and peer-to-peer exchange)
from a dozen of laboratories.


 Examples
of
Neuronal Reconstructions
Examples
of
Neuronal Reconstructions
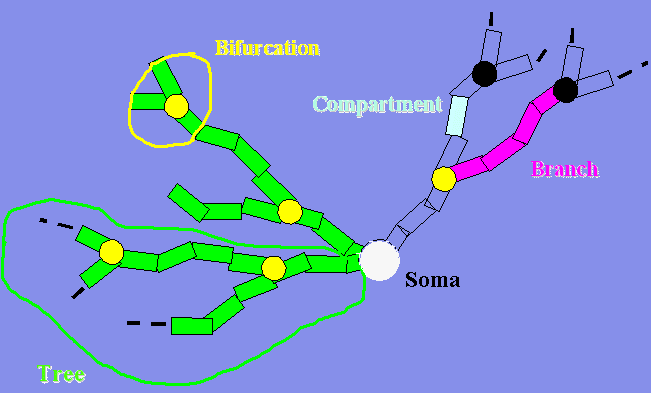
The process of extraction of morphological measurements with L-Measure
can be logically divided in sequential steps, corresponding to separate
panels in the graphical user interface:
1) The files corresponding to one or more neurons are selected from
the local drive (Input panel)
2) The morphometric function or group of functions is selected,
defining what will be measured (Function panel)
3) The part of the tree to be analyzed (region of interest) is
selected (Specificity panel)
4) The format of the output is selected, specifying the name of the
file to be saved, etc. (Output panel)
5) The analysis is launched (Go panel)
6) Search is carried out on a file or a group of files considering different statistical functions
(LMSearch Panel)
Steps 3, 4 and 6 are optional (i.e., an analysis can be run with just
1-2-5). If no part of the neuron is specified (step 3 is skipped), the
measurement will be extracted from the whole neuron. If the format of
the output is not specified (step 4 is skipped), the result will be
displayed as ASCII text in the Go panel. If the search is not required,
step 6 can be skipped
For questions or comments, please contact Sridevi Polavaram