Width
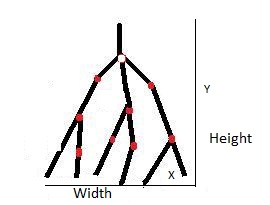
Width is computed on the x-coordinates
and it is the difference of minimum and maximum x-values after
eliminating the outer points on the either ends by using the 95%
approximation of the x-values of the given input neuron.
Function Output Type : Real
Calculated : At each tracing point
Returns a value : For whole arbor
Output:
| Metric | Total_Sum | #Compartments (considered) |
#Compartments (discarded) |
Minimum | Average | Maximum | S.D. |
| Soma_Surface | 159.722 | 1 | (1101) | 159.722 | 159.722 | 159.722 | 0 |
Values to consider : Min/ Avg/ Max
Output Interpretation :
If the user believes that the input neuron is not oriented properly, one can do principal component analysis (PCA) first by selecting PCA in the function panel. In this case, width is computed using the same 95% approximation on the new x-axis after shifting and rotating the cell to a different axis based on component analysis.
Total_Sum = 159.722 gives Total_Sum, Minimum = 159.722 gives
minimum, Maximum = 159.722 gives maximum for given input neuron.
References :

- Although the X-value at each tracing point is considered, the width is computed for the entire Neuron. So, one should only use the Min/Avg/Max values for this metric. NOT Total_Sum.
-One can choose the rule "type=3" in the specificity panel to limit the width analysis to dendrites only.
-One can choose the rule "type=3" in the specificity panel to limit the width analysis to dendrites only.